Ministry of Science & Technology
New method for detecting H. pylori & its mutations can help dyspeptic patients in resource poor remote settings
Posted On:
27 DEC 2024 3:13PM by PIB Delhi
Researchers have found a way to develop FELUDA as a point-of-care diagnostic service at a minimal cost for detection of H. pylori and its mutations in dyspeptic patients from rural areas of India, with minimal or no access to diagnostic laboratories.
Infections with H. pylori affect over 43 percent of the world’s population with a wide range of gastrointestinal disorders, including peptic ulcers, gastritis, dyspepsia and even gastric cancer.
Resistance to clarithromycin, primarily attributed to point mutations in the 23S ribosomal RNA coding gene of H. pylori poses a global threat to public health, by necessitating repeated diagnostic tests and use of multiple courses of different antibiotic combinations for eradication of the same.
Therefore, integration of novel diagnostic strategies as cost-effective diagnostic tools to detect the presence of H. pylori in human samples, as well as the identification of the antibiotic susceptibility is crucial for its rapid eradication.
CRISPR-based methodologies are known to enable site recognition and cleavage of the target DNA with exceptional accuracy by designing guide RNAs targeting the respective mutation site in various kinds of DNA samples. Hence, in-depth understanding of H. pylori genetic makeup by CRISPR-based diagnostics (CRISPRDx) could aid in molecular dissection of its pathogenicity and development of targeted therapeutics against different strains.
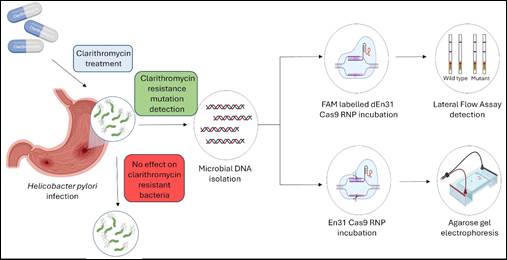
Towards this goal, Dr. Debojyoti Chakraborty and Dr. Souvik Maiti’s group at CSIR-IGIB had previously demonstrated the possibility of detecting H. pylori antibiotic resistance mutations using Cas9-based mutation detection strategies. However, CRISPR-Cas9 based biosensing techniques face limitations due to the requirement of NGG PAM sequences at the recognition site while detecting mutations.
To encounter this limitation of CRISPR-Cas9 based detection tools in this study, Dr Shraddha Chakraborty (currently a Department of Science and Technology INSPIRE Faculty Fellow at DBEB, IIT Delhi) and colleagues at CSIR-IGIB explored the potential of en31-FnCas9 to successfully detect the presence and identify the 23S rDNA mutation status of H. pylori in gastric biopsy samples from dyspeptic patients, both by in vitro cleavage studies and lateral flow-based test strip assays (FELUDA). Clinical arm of the study was led by Dr. Govind K. Makharia (Department of Gastroenterology, AIIMS New Delhi), Dr. Manas K. Panigrahi (Department of Gastroenterology, AIIMS Bhubaneswar) and Dr. Vinay K. Hallur (Department of Microbiology, AIIMS Bhubaneswar).
They used an engineered Cas9 protein having resemblance to Cas9 orthologs isolated from Francisella novicida (en31-FnCas9) but with altered PAM binding affinity.
In their paper published in the Microchemical Journal they reported the potential of this en31-FnCas9 to successfully detect the presence and identify the 23S rDNA mutation status of H. pylori in gastric biopsy samples from dyspeptic patients of Indian origin.
The study highlights the significance of sequencing-free molecular diagnosis in detecting H. pylori and its antibiotic resistance mutations, thereby emphasizing the need for tailored treatment plans to address global public health concerns associated with antibiotic resistance and gastric cancer risks.
The integration of en31-FnCas9-based detection with lateral flow assay (FELUDA) demonstrated rapid visual readout of H. pylori infection and its mutation status in patient samples, enhancing its diagnostic potential in clinical settings.
This is the first report of en31-FnCas9 mediated molecular diagnosis of H. pylori mutations implicated in clarithromycin resistance.
Successful deployment this methodology in a clinical setup can be helpful in providing accurate and timely reports on the antibiotic resistance pattern of the H. pylori strains isolated from patients, in remote settings allowing for effective management of this global public health concern.
***
NKR/KS
(Release ID: 2088376)
Visitor Counter : 1592