Ministry of Science & Technology
Indian scientists helped rewrite a 50-Year-Old biological rule
Posted On:
18 NOV 2025 5:07PM by PIB Delhi
A new study overturns a central textbook model of bacterial gene regulation and unveils new paths for understanding bacterial gene regulation and its evolution. This can help in designing better antibiotics or regulatory inhibitors that block infection mechanisms.
The understanding of how bacteria control their genes, affects our concept of everything from how microbes respond to stress to how we design antibiotics that stop them in their tracks. If the basic mechanics of gene regulation differ between species, it could mean new strategies for fighting infections, or even for harnessing bacteria to produce useful compounds.
For nearly 50 years, biology has related the story of how bacteria turn their genes on with the help of the so-called “σ (sigma) cycle” – factors that bind RNA polymerase to initiate transcription and then dissociate to allow elongation. This concept was built largely on observations of bacterial strain E. coli σ70.
However, Researchers from the Bose Institute, an autonomous institute of the Department of Science and Technology (DST) and Rutgers University reveal that the cycle is not a universal phenomenon.
In a study published in the Proceedings of the National Academy of Sciences (PNAS) they have reported that, contrary to decades of scientific belief, the principal transcription initiation factor in Bacillus subtilis—σA—and a modified version of the Escherichia coli σ70 factor remain bound to RNA polymerase throughout transcription, rather than being released after initiation.
“Our work shows that in Bacillus subtilis, the σA factor stays attached to RNA polymerase all the way through the transcription process,” said Dr. Jayanta Mukhopadhyay, corresponding author from the Bose Institute. “This fundamentally changes how we think about bacterial transcription and gene regulation,” he added.
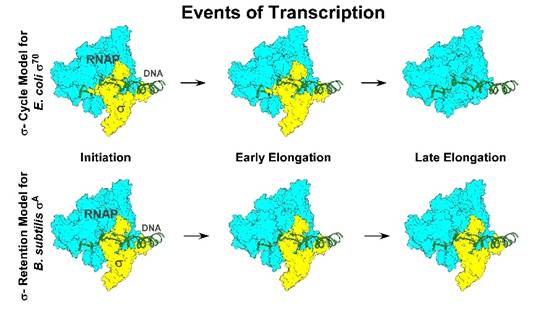
Using a combination of modern techniques like biochemical assays, chromatin immunoprecipitation, and fluorescence-based imaging — the researchers watched the sigma factor’s behaviour in real time. They found that Bacillus subtilis σA and an E. coli σ70 variant lacking a part called 1.1 remain stably associated with transcription complexes. This is in stark contrast to full-length E. coli σ70, which is released stochastically during elongation.
“These findings provide compelling evidence that the long-accepted σ cycle does not apply to all bacteria,” added co-author Aniruddha Tewari of Bose Institute. “It opens new avenues for understanding bacterial gene regulation and its evolution.”
The discovery has broad implications for microbiology, potentially influencing how researchers approach bacterial physiology, stress response, and the development of antibiotics targeting transcription.
By controlling gene regulation, scientists can design microorganisms that produce biofuels, biodegradable plastics, or therapeutic compounds efficiently.
Besides, Dr Tewary and Dr Mukhopadhyay, Shreya Sengupta, Soumya Mukherjee, Nilanjana Hazra from Bose Institute, Kolkata and Yon W. Ebright, Richard H. Ebright from Rutgers University, USA contributed in the study.
Publication link: doi:10.1073/pnas. 2503801122
*****
NKR/AK
(Release ID: 2191262)
Visitor Counter : 1828